Hyperparameter Tuning for Neural Networks with Ray Tune
Author : Emre Okcular
Date : June 7st 2021

In this post, we will take a closer look at hyperparameters of a deep neural network listed below. Moreover, we will cover how to tune these parameters similar approach with scikit-learn GridSearch. Ray Tune package is a Python library for experiment execution and hyperparameter tuning at any scale.
Important hyperparameters for Neural Networks:
- Learning Rate
- Dropout
- Weight Decay
- Batch Size
- Number of Epochs
Learning Rate
Learnin rate is the most important hyper-parameter to tune for training neural networks. If learning rates are too small it causes slow training, if learning rates are too large is causes training to diverge. As an initial approach we can fix the learning rate and then monotonically decrease during training.
Dropout
Dropout is a regularization technique that prevents overfitting in neural networks. At each training stage (batch), individual nodes are either dropped out of the net with probability p or kept with probability 1-p, so that a reduced network is left.
Training Phase
For each hidden layer, for each training sample, for each iteration, ignore (zero out) a random fraction, p, of nodes (and corresponding activations).
Testing Phase
Use all activations, but reduce them by a factor 1-p (to account for the missing activations during training).
Neural networks learn co-adaptations of hidden units that work for the training data but do not generalize to unseen data.Random dropout breaks up co-adaptations making the presence of any particular hidden unit unreliable.
Weight Decay
It is usually a parameter in the optimization function such as torch.optim.Adam(params, lr=0.001, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=False) . In other words it is a L2 penalty that is added to error function. Larger values of λ will tend to shrink the weights toward zero: typically cross-validation is used to estimate λ.
Batch Size
Batch size is a number of samples processed before the model is updated.
Why to use minibatch?
- How much data can you fit on your GPU.
- Small batches can offer a regularizing effect, possibly due to the noise they add to the learning process.
- The model update frequency is higher with mini-batches.
- Larger batches provide more accurate estimate of the gradient.
Number of Epochs
Number of epochs is the number of complete passes through the training dataset.
While tuning these parameters manually, you might feel like a gradient decent in multidimensional space looking for some pattern(increase or decrease). In scikit-learn pipelines we use methods like HalvingGridSearchCV , RandomizedSearchCV , GridSearchCV to solve this search problem. It may take some time and CPU heat with big datasets but eventually you will have tuned model with best estimator and hyperparameters. With the same approach we can tune neural network parameters using Ray Tune package in efficient and distributed way.
We will import below packages from PyTorch and Ray.
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader, WeightedRandomSampler
from ray import tune # For hyperparameter tuning in Neural Networks.
from ray.tune import Analysis # For analyzing tuning results.
Let use a simple NN model architecture with one layer, dropout, ReLU activation function and batchnorm.
Model Architecture
class MulticlassClassification(nn.Module):
def __init__(self, num_feature, num_class,d,M):
super(MulticlassClassification, self).__init__()
self.layer_1 = nn.Linear(num_feature, M)
self.layer_out = nn.Linear(M, num_class)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(p=d)
self.batchnorm1 = nn.BatchNorm1d(M)
def forward(self, x):
x = self.layer_1(x)
x = self.batchnorm1(x)
x = self.relu(x)
x = self.dropout(x)
x = self.layer_out(x)
return x
To use tuning method we need to convert our training code to a function and add tune.report(training_accuracy=train_epoch_acc/len(train_loader),val_accuracy=val_epoch_acc/len(val_loader),training_loss=train_epoch_loss/len(train_loader),val_loss=val_epoch_loss/len(val_loader) to our training loop to store the metrics for each epoch.
def train(model,optimizer,train_loader,val_loader,EPOCHS,accuracy_stats,loss_stats):
for e in tqdm(range(1, EPOCHS+1)):
train_epoch_loss = 0
train_epoch_acc = 0
model.train()
for X_train_batch, y_train_batch in train_loader:
X_train_batch, y_train_batch = X_train_batch, y_train_batch
optimizer.zero_grad()
y_train_pred = model(X_train_batch)
train_loss = criterion(y_train_pred, y_train_batch)
train_acc = multi_acc(y_train_pred, y_train_batch)
train_loss.backward()
optimizer.step()
train_epoch_loss += train_loss.item()
train_epoch_acc += train_acc.item()
# VALIDATION
with torch.no_grad():
val_epoch_loss = 0
val_epoch_acc = 0
model.eval()
for X_val_batch, y_val_batch in val_loader:
X_val_batch, y_val_batch = X_val_batch, y_val_batch
y_val_pred = model(X_val_batch)
val_loss = criterion(y_val_pred, y_val_batch)
val_acc = multi_acc(y_val_pred, y_val_batch)
val_epoch_loss += val_loss.item()
val_epoch_acc += val_acc.item()
loss_stats['train'].append(train_epoch_loss/len(train_loader))
loss_stats['val'].append(val_epoch_loss/len(val_loader))
accuracy_stats['train'].append(train_epoch_acc/len(train_loader))
accuracy_stats['val'].append(val_epoch_acc/len(val_loader))
tune.report(training_accuracy=train_epoch_acc/len(train_loader),val_accuracy=val_epoch_acc/len(val_loader),training_loss=train_epoch_loss/len(train_loader),val_loss=val_epoch_loss/len(val_loader))
return accuracy_stats
def get_loaders():
df = pd.read_csv("/Users/emre/Dev/GitHub/train_ml2_2021.csv")
y=df['target']
X=df.drop(columns='target')
test = pd.read_csv('/Users/emre/Dev/GitHub/test0.csv')
X_test = test.drop(columns=['target',"obs_id"])
y_test = test["target"]
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.2, stratify=y, random_state=21)
scaler = MinMaxScaler()
X_train = scaler.fit_transform(X_train)
X_val = scaler.transform(X_val)
X_test = scaler.transform(X_test)
X_train, y_train = np.array(X_train), np.array(y_train)
X_val, y_val = np.array(X_val), np.array(y_val)
X_test, y_test = np.array(X_test), np.array(y_test)
train_dataset = CancerDataset(torch.from_numpy(X_train).float(), torch.from_numpy(y_train).long())
val_dataset = CancerDataset(torch.from_numpy(X_val).float(), torch.from_numpy(y_val).long())
test_dataset = CancerDataset(torch.from_numpy(X_test).float(), torch.from_numpy(y_test).long())
target_list = []
for _, t in train_dataset:
target_list.append(t)
target_list = torch.tensor(target_list)
target_list = target_list[torch.randperm(len(target_list))]
c = Counter(y_train)
class_count = [i for i in [i[1] for i in sorted(c.items())]]
class_weights = 1./torch.tensor(class_count, dtype=torch.float)
class_weights_all = class_weights[target_list]
weighted_sampler = WeightedRandomSampler(
weights=class_weights_all,
num_samples=len(class_weights_all),
replacement=True)
return weighted_sampler, train_dataset, val_dataset,len(X.columns),y.nunique()
Finally, we will use train function to wrap all the helpers.
def train(config):
test = []
accuracy_stats = {
'train': [],
"val": []
}
loss_stats = {
'train': [],
"val": []
}
w,tr,val,num_feat, num_cl = get_loaders()
train_loader = DataLoader(dataset=tr,
batch_size=config["bs"],
sampler=w, drop_last=True)
val_loader = DataLoader(dataset=val,shuffle=True, batch_size=1)
model = MulticlassClassification(num_feature = NUM_FEATURES, num_class=NUM_CLASSES,d=config["dr"],M=config["m"])
optimizer = optim.Adam(model.parameters(), lr=config["lr"],weight_decay = config["wd"])
train(model, optimizer, train_loader, val_loader,EPOCHS=EPOCH_FOR_TUNING,accuracy_stats=accuracy_stats,loss_stats=loss_stats) ##EPOCH is hardcoded!
Before hyperparameter search we need to specify the search space in a dictionary.
param_space = {"lr": tune.grid_search([0.00003,0.0005, 0.001, 0.0007]), # Learning Rate
"bs": tune.grid_search([8, 16, 32,64, 128 , 256,512]), # Batch Size
"dr": tune.grid_search([0.3, 0.5,0.7,0.85]), # Dropout
"m": tune.grid_search([30, 50, 100, 200, 300]), # Number of neurons
"wd": tune.grid_search([0])} # Weight Decay for optimizer
When we run below method it will start to search parameter space by creatgin folders in the root library.
analysis = tune.run(train, config=param_space,verbose=1, name="epoch_"+str(EPOCH_FOR_TUNING))
You can see the folders in the path /Users/emre/ray_results/epoch_300 Here epoch_300 is the name for tuning process we specified above in the run() method.
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00000_0_bs=8,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00001_1_bs=16,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00002_2_bs=32,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00003_3_bs=64,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00004_4_bs=128,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00005_5_bs=256,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00007_7_bs=8,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00008_8_bs=16,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00009_9_bs=32,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-08
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00010_10_bs=64,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-35
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00011_11_bs=128,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-35
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00012_12_bs=256,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-36
drwxr-xr-x 8 emre staff 256 6 May 11:35 train_cancer_c3378_00013_13_bs=512,dr=0.5,lr=3e-05,m=30,wd=0_2021-05-06_11-35-41
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00014_14_bs=8,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-35-47
drwxr-xr-x 7 emre staff 224 6 May 11:35 train_cancer_c3378_00015_15_bs=16,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-35-53
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00016_16_bs=32,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-35-53
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00017_17_bs=64,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-36-05
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00018_18_bs=128,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-36-07
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00019_19_bs=256,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-36-19
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00021_21_bs=8,dr=0.85,lr=3e-05,m=30,wd=0_2021-05-06_11-36-29
drwxr-xr-x 8 emre staff 256 6 May 11:36 train_cancer_c3378_00020_20_bs=512,dr=0.7,lr=3e-05,m=30,wd=0_2021-05-06_11-36-29
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00022_22_bs=16,dr=0.85,lr=3e-05,m=30,wd=0_2021-05-06_11-36-31
drwxr-xr-x 7 emre staff 224 6 May 11:36 train_cancer_c3378_00023_23_bs=32,dr=0.85,lr=3e-05,m=30,wd=0_2021-05-06_11-36-37
drwxr-xr-x 2 emre staff 64 6 May 11:36 train_cancer_c3378_00024_24_bs=64,dr=0.85,lr=3e-05,m=30,wd=0_2021-05-06_11-36-56
-rw-r--r-- 1 emre staff 2108895 6 May 11:36 experiment_state-2021-05-06_11-34-59.json
-rw-r--r-- 1 emre staff 6215 6 May 11:36 basic-variant-state-2021-05-06_11-34-59.json
drwxr-xr-x 8 emre staff 256 6 May 17:52 train_cancer_c3378_00006_6_bs=512,dr=0.3,lr=3e-05,m=30,wd=0_2021-05-06_11-35-01
Lets take a look at one of the folder.
-rw-r--r-- 1 emre staff 63 6 May 11:35 params.json
-rw-r--r-- 1 emre staff 64 6 May 11:35 params.pkl
-rw-r--r-- 1 emre staff 80366 6 May 11:36 progress.csv
-rw-r--r-- 1 emre staff 208199 6 May 11:36 result.json
-rw-r--r-- 1 emre staff 169618 6 May 11:36 events.out.tfevents.1620326101.Emre-MacBook-Pro.local
You can find NN parameters, weights, model as pkl object, and logs.
print("Best config: ", analysis.get_best_config(mode ="max" ,metric="val_accuracy"))
Best config: {'lr': 0.0005, 'bs': 128, 'dr': 0.3, 'm': 300, 'wd': 0}
Saving Results as DataFrame
| training_accuracy | val_accuracy | training_loss | val_loss | time_this_iter_s | done | timesteps_total | episodes_total | training_iteration | experiment_id | date | timestamp | time_total_s | pid | hostname | node_ip | time_since_restore | timesteps_since_restore | iterations_since_restore | trial_id |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 45.24390243902439 | 40.54054054054054 | 0.7499703270633046 | 0.7465202832544172 | 3.6098971366882324 | False | 1 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-08 | 1620326108 | 3.6098971366882324 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 3.6098971366882324 | 0 | 1 | c3378_00000 | ||
| 49.97560975609756 | 35.13513513513514 | 0.7230090389891368 | 0.7733307557331549 | 0.1837749481201172 | False | 2 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-08 | 1620326108 | 3.7936720848083496 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 3.7936720848083496 | 0 | 2 | c3378_00000 | ||
| 48.19512195121951 | 35.13513513513514 | 0.7115627977906204 | 0.7763204888717549 | 0.1500110626220703 | False | 3 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-08 | 1620326108 | 3.94368314743042 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 3.94368314743042 | 0 | 3 | c3378_00000 | ||
| 54.829268292682926 | 37.83783783783784 | 0.6684811834881945 | 0.7417761256565919 | 0.17262482643127441 | False | 4 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-09 | 1620326109 | 4.116307973861694 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 4.116307973861694 | 0 | 4 | c3378_00000 | ||
| 52.41463414634146 | 40.54054054054054 | 0.6830459309787285 | 0.7908904548432376 | 0.11512398719787598 | False | 5 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-09 | 1620326109 | 4.23143196105957 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 4.23143196105957 | 0 | 5 | c3378_00000 | ||
| 56.63414634146341 | 45.945945945945944 | 0.6508946157083279 | 0.7613682513301437 | 0.13743996620178223 | False | 6 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-09 | 1620326109 | 4.3688719272613525 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 4.3688719272613525 | 0 | 6 | c3378_00000 | ||
| 59.73170731707317 | 45.945945945945944 | 0.62467926449892 | 0.7794352487937825 | 0.14998722076416016 | False | 7 | 55bb3e915a12438a9574e1cecc1423dd | 2021-05-06_11-35-09 | 1620326109 | 4.518859148025513 | 40040 | Emre-MacBook-Pro.local | 192.168.0.155 | 4.518859148025513 | 0 | 7 | c3378_00000 |
You can save the results as a dataframe.
analysis.dataframe().to_csv("df_ep_100.csv")
For example, you can draw accuracy and loss graphs using this data.
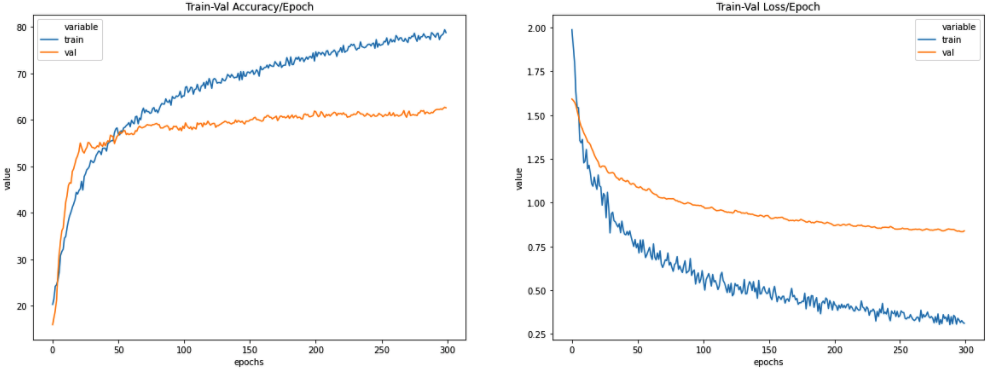
Loading old Results
If you want to take a look at previous tuning analysis you can load with below method.
analysis2 = Analysis("/Users/emre/ray_results/train_cancer_2021-05-01_22-32-12")
Conclusion
To wrap up, hyperparameter tuning in neural networks is an exhaustive search with lots of time and CPU power. Be careful with the parameter space that you used. It may give you the best parameters in somewhere in the parameter space which are not related with the exact solution.
Thank you for reading.